|
Data Sources |
| Getting Started |
|
The Statistics |
|
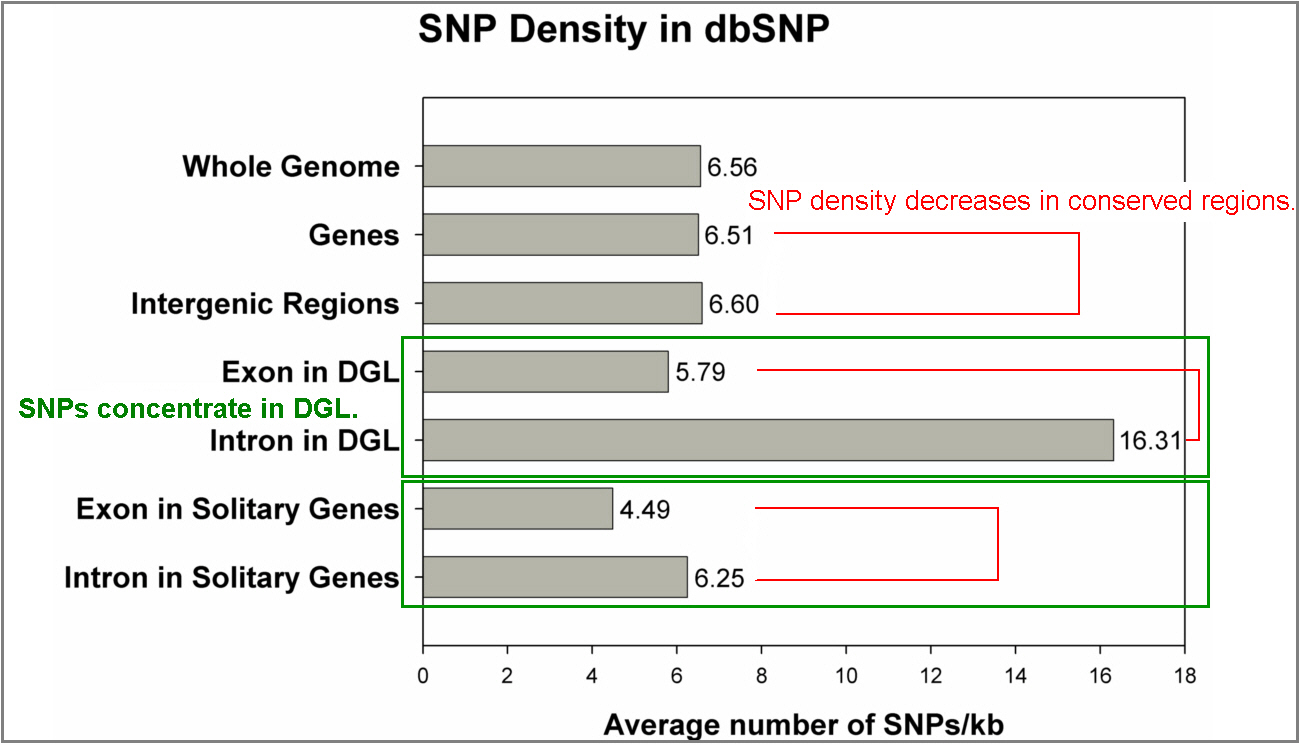
SNP density decreases in more conserved regions, such as exons. This may reflect that functional regions are more conserved under the pressure of selection during evolution. Thus, they are not prone to maintaining diversity. |
|
The Distribution of DNV and DNV-coupled SNPs 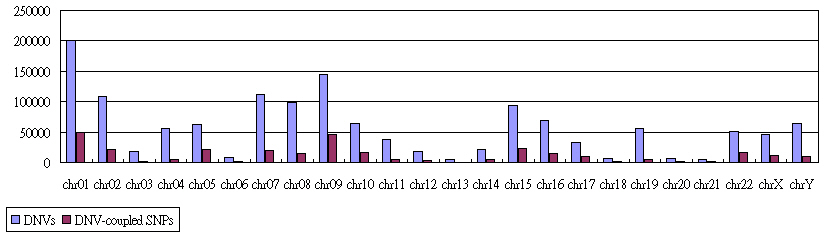
|
|
Counts of SNPs, DNVs and DNV-coupled SNPs in Duplicated Genes 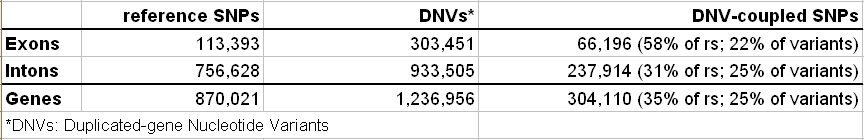
|