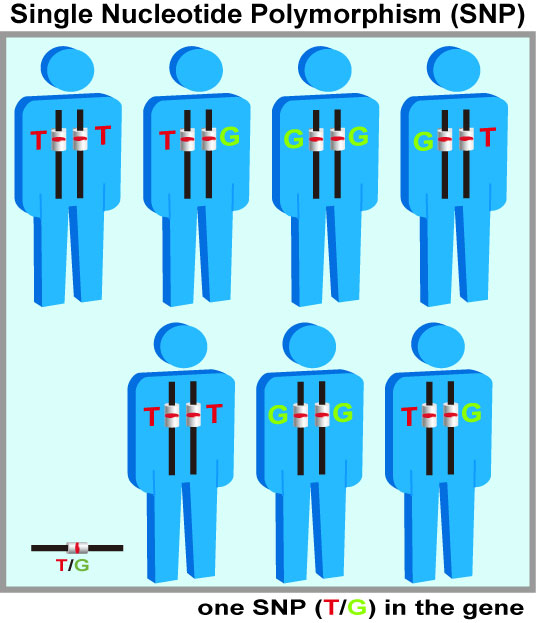
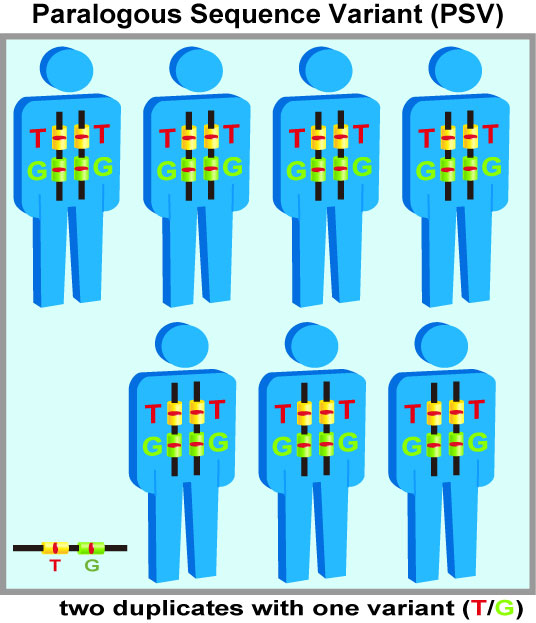
Gene duplications are scattered widely throughout the human genome. A single-base difference located in nearly identical duplicated segments may be misjudged as a single nucleotide polymorphism (SNP) from individuals (right figures). A lack of detailed clarification of these undistinguishable SNPs located in duplicates can inflate the heterozygosity of SNPs. These ambiguous SNP calls are observed under current genotyping methods using assays of PCR, short DNA sequencing, and hybridization to DNA microarrays or beads. This confounding phenomenon is even more serious when identifying SNPs and mutations by shorter DNA fragments randomly produced from the next-generation sequencing (NGS). As the next-generation sequencing become more popular for sequence-based association studies, numerous ambiguous SNPs are rapidly accumulated.
The dbDNV is established to promote more accurate variation annotations. We have identified over 10% of human genes associated with duplicated gene loci (DGL). Through sequence alignments of DGL, we systematically designated 1,236,956 variations as duplicated-gene nucleotide variants (DNVs). The dbDNV contains 304,110 DNV-coupled SNPs which cover approximately 58% of exonic SNP records in DGL. Because of high accumulation of ambiguous SNPs, we suggest that annotating SNPs with DNVs possibilities should improve association studies of these variants with human diseases.
Publication
“dbDNV: a resource of duplicated-gene nucleotide variants in human genome,” Meng-Ru Ho, Kuo-Wang Tsai, Chun-houh Chen, and Wen-chang Lin, Nucleic Acids Research, 2011 January; Volume 39 (suppl 1): D920 – D925.
References
1. "Complex SNP-related sequence variation in segmental genome duplications," Fredman D et al. (2004) Nature Genetics 36, p. 861–866.
2. "Duplicating SNPs," Gut IG and Lathrop GM (2004) Nature Genetics 36, p. 789–790.