There are three search functions established in the dbDNV. |
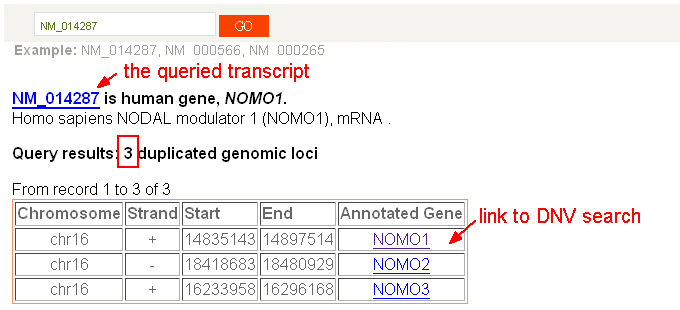 |
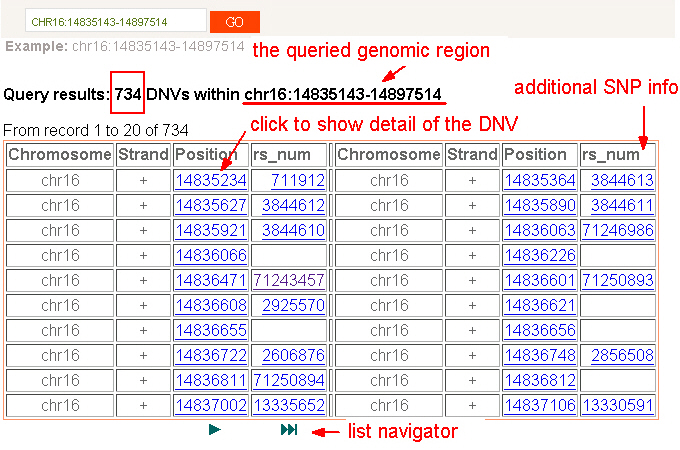
2) DNVs within a genomic region. Users can browse the identified DNVs in the genomic order by clicking the chromosome name. In addition to browsing by chromosomes, users can get a list of DNVs after specifying a chromosome or a genomic region. The dbDNV annotates each DNV by its 50-bases flanking sequence. UUsers then can explore the related duplicating segments that generate the specified DNV by clicking the DNV’s position. The multiple sequence alignment of listed duplicating segments could demonstrate the interfering capability of the DNV via mimicking SNP calls in genotyping. Moreover, users can extend their search by click the DNV-related transcripts of the queried DNV. Furthermore, some DNVs may possess existing SNP records in dbSNP. If users click “rs_num” instead of “position”, the dbDNV displays the genotype of the annotated SNP record accordingly. Users can also link out to dbSNP for detailed information. [GO] |
 |
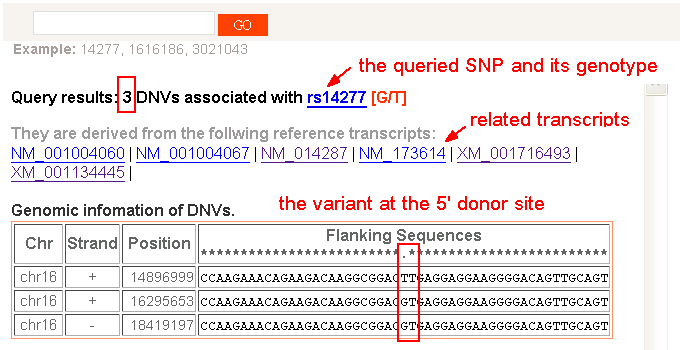
3) DNV-coupled SNPs. This is the search page for looking at existing SNP records in dbSNP that are also identified as DNVs. Users can explore the DNV located on the same genomic position of the queried SNP. The related transcripts utilized to identify the DNV are listed. These transcripts are linked to the DGL search. Thus, users can use a preferred SNP record to pull down the comprehensive set of paralogous loci and their DNVs. In addition to the annotated genotype of the queried SNP, the variant’s flanking sequences in multiple duplicated loci are shown. [GO] |
 |
The dbDNV annotates DNVs with affiliated paralogous segments and existing SNP records in dbSNP. The first search function is mainly employed to display the duplication structure of queried genes. The second one can provide DNV annotation within a user specified region. By manipulating these two search functions together, users explore the gene duplication structure and the associated DNVs. The dbDNV annotates DNVs with affiliated paralogous segments and existing SNP records in dbSNP. Users can perform in-depth investigation if necessary. This promotes more accurate SNP annotation. The third search function is designed for examining existing SNP records with possibility of DNVs. As controversial SNP records have been accumulated in public, SNPs may require the inspection of existing in duplications before being further inferred.
Aside from providing the search function, dbDNV also offers all users a flat file download without any restriction. Users can establish their own global genome examination system in genetic analysis on the basis of our DNV annotation. Please visit the download page.